varDB: Antigenic variation database
Many phylogenetically diverse organisms use antigenic variation to avoid recognition by the immune system.
varDB was developed to serve as a centralized database of antigenically variable protein families from a range of pathogenic organisms.
Infectious diseases and their pathogens are often studied in isolation, but observations made in one species may be used to formulate hypotheses relevant to other taxa.
VarDB is intended to facilitate large-scale examination of the evolution of antigenic gene families
and help provide insight into how a conserved function can be maintained despite extreme sequence diversity.
|
Sequences by country: 1
1,309
|
Isolates by country Number of GenBank antigenic variation field isolate sequences by country (extracted from the GenBank country field). |
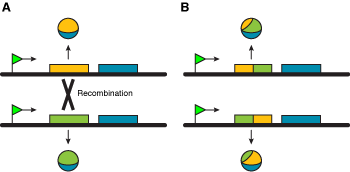 |
Non-homologous recombination. The two chromosomes in (A) encode different polypeptides. A recombination event ocurring in the first exon allows the generation of diversity (B). |
Acknowledgments
This website was developed as part of a collaboration between
Kyoto University in Japan
and the Karolinska Institute in Sweden.
We are grateful to the STINT Foundation, the Swedish Royal Academy of Sciences, the Japan Society for the Promotion of Science, and the Ministry of Education, Culture, Sports, Science and Technology of Japan for funding.
If you use varDB in your work, please cite the following publication:
Hayes CN, Diez D, Joannin N, Honda W, Kanehisa M, Wahlgren M, Wheelock CE, Goto S. 2008. varDB: a pathogen-specific sequence database of protein families involved in antigenic variation. Bioinformatics (Oxford, England) 24:2564-5. (18776192)
Bioinformatics Center,
Institute for Chemical Research,
Kyoto University, Uji, Kyoto 611-0011, Japan
System last updated Thu, January 24, 2013 14:25:29 JST / SVN revision: 1054
Username: 764F73F90BAE31298C2D6E1C3E1F9E3A
